Coronavirus Structural Task Force
As methods developers in structural biology, we usually work far from the spotlight. When the COVID-19 pandemic began, we asked ourselves: How can we contribute to the fight against the virus?

As early as February 2020, we started evaluating the structures of macromolecules in SARS-CoV and later SARS-CoV-2 available from the Protein Data Bank and found ample room for improvement. We set up a website (https://insidecorona.net) and a database containing our evaluation and revised models. We have established an automatic structure evaluation and revise individual structures on a weekly basis. We also engage in outreach activities, writing blog posts about the structural biology of SARS-CoV-2 aimed at both the scientific community and the general public, refining structures live on Twitch and offering a 3D-printable virus model for schools.
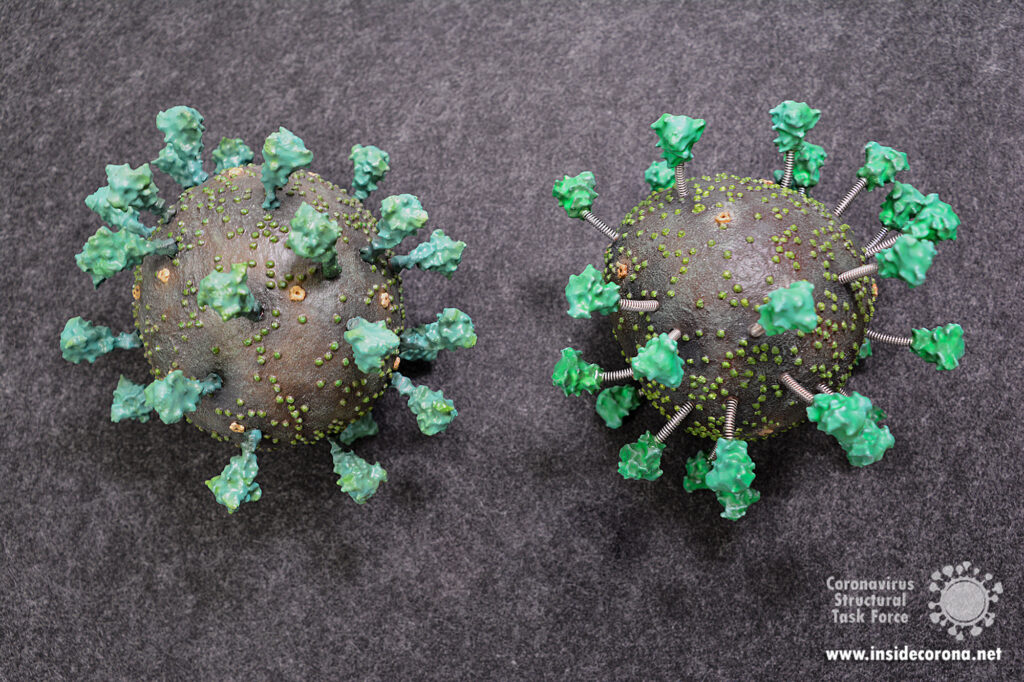
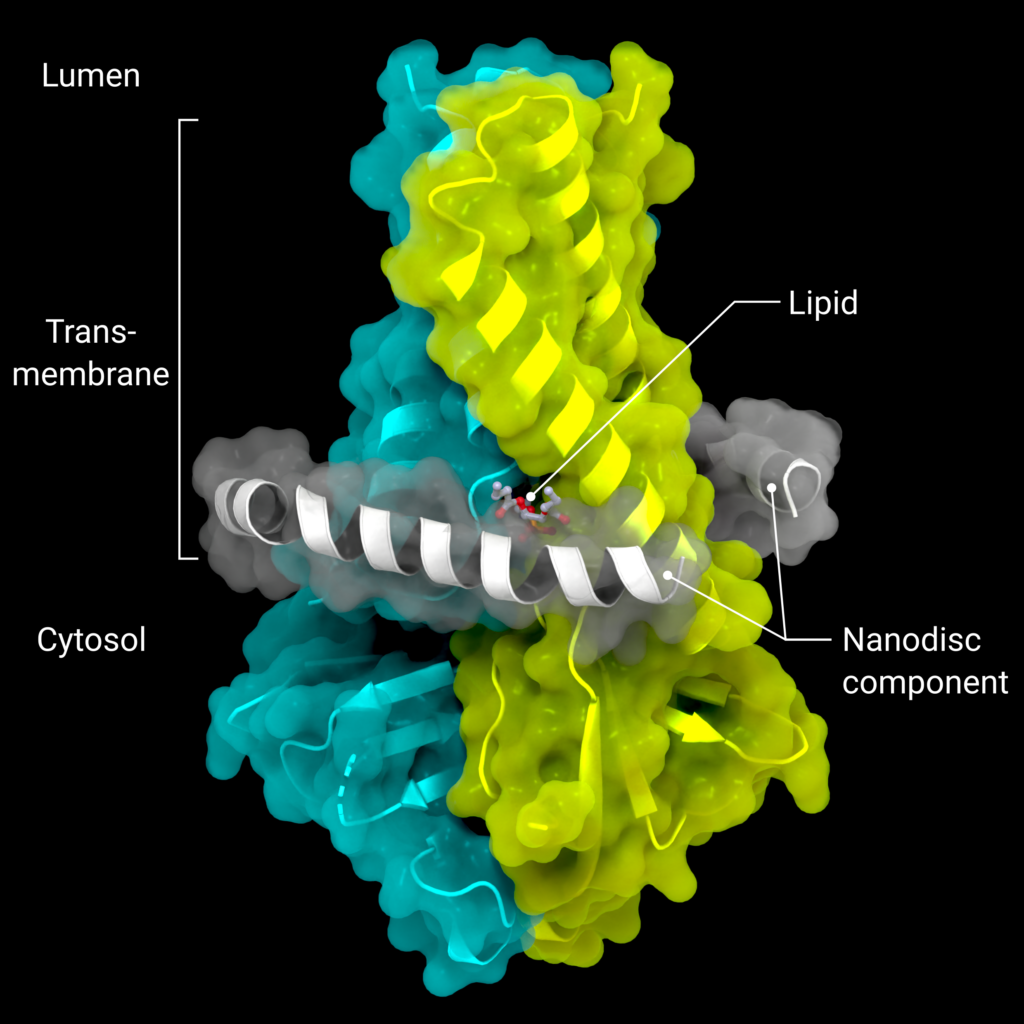
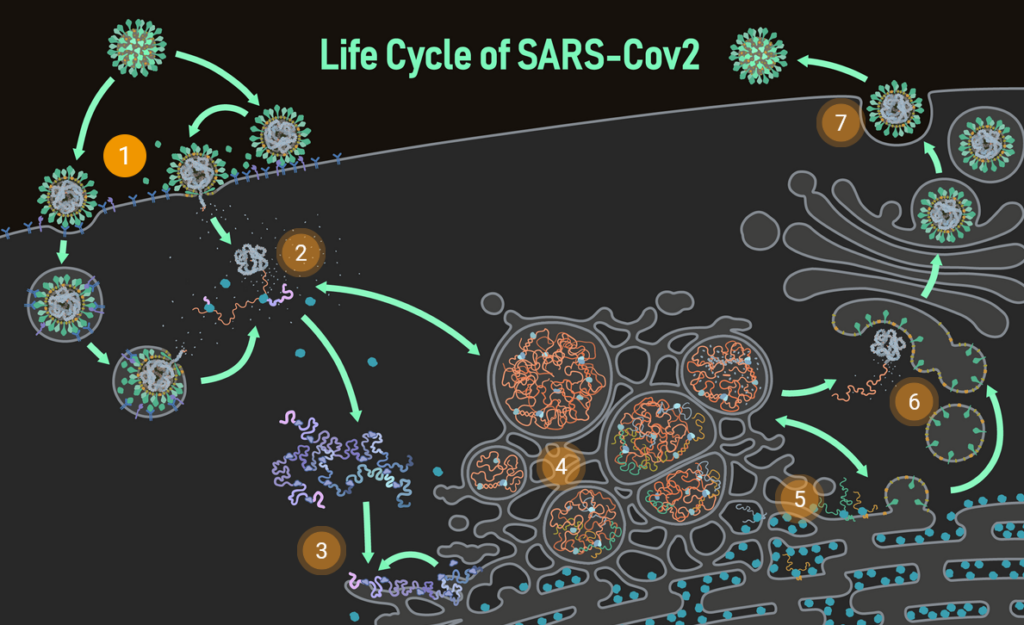
In the beginning, there were no tenured academics in the Coronavirus Structural Task Force; we were an ad hoc collaboration of mostly junior researchers across nine time zones, brought together by the desire to fight the pandemic [1]. We lacked management experience, computing facilities and administrative infrastructure. Still, we were able to rapidly establish a large network of COVID-19-related research, forge friendships and collaborations across national boundaries, spread knowledge about the structural biology of the virus and provide improved models for in-silico drug discovery projects. We may have gotten into the spotlight after al
[1] Croll, T., Diederichs, K., Fischer, F., Fyfe, C., Gao, Y., Horrell, S., Joseph, A. P., Kandler, L., Kippes, O., Kirsten, F., Müller, K., Nolte, K., Payne, A., Reeves, M. G., Richardson, J., Santoni, G., Stäb, S., Tronrud, D., Williams, C. & Thorn, A. (2020). BioRxiv. doi:10.1101/2020.10.07.307546.