Cryo-EM
Modelling Large Complexes in Cryo Electron Microscopy
Cryo-EM is a rapidly developing method, which is providing unprecedented structural information about large molecular machines, membrane complexes, and especially interaction between proteins. However, the computational methods needed to analyse the data coming out of this exciting new field currently lag well behind what is experimentally possible. Indeed, most tools used to interpret reconstruction maps were originally developed for crystallography. While these tools are able to deliver reasonable static or average structures, they do not take full advantage of the wealth of biological information available from the experimental data, nor are they entirely well-suited to modelling cryo-EM data. As a consequence, we throw away information that could provide novel biological insights and manual map interpretation remains very challenging, in particular if the structures being modelled require de novo model building or are very mobile. This challenge must be urgently addressed by the development not only of new models, but also by new tools that are specifically designed to fit the nature of the sample and the cryo-EM experiment. One such tool I have already developed is Haruspex, a convolutional neural network that can be straightforwardly applied to newly reconstructed maps in order to support domain placement or as a starting point for main chain placement.
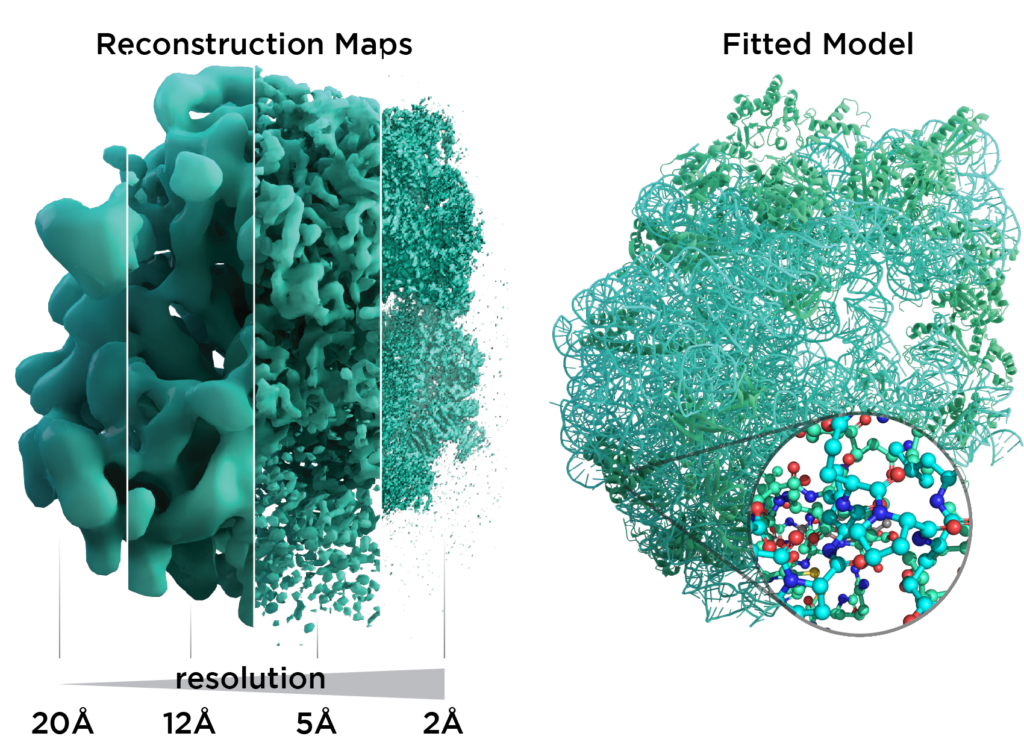
Resolution revolution in Cryo-EM. Left: Reconstruction maps shown for the 50S ribosome with increasing level of detail from left to right as the field evolved. Right: A typical atomic model used for interpretation of reconstruction maps. Picture by scistyle.com
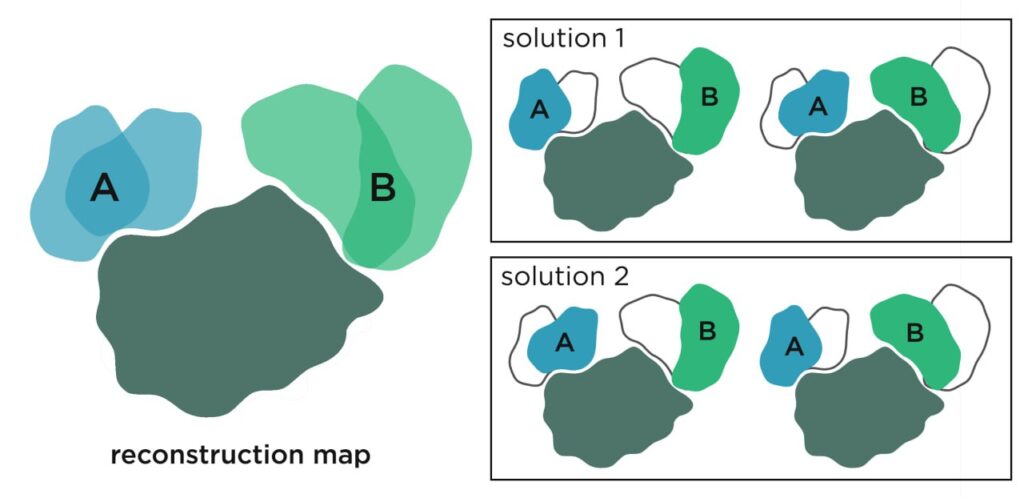
Coordinated movement information is lost in 3D reconstruction maps. A reconstruction map could be resulting from different coordinated movement (solution 1 or 2) – or from no coordinated movement at all, given the ambiguous reconstruction map (left), while the individual micrographs would contain this information. Currently, this can only be resolved if the data set is large, the movement simple and the states discrete by solving two distinct structures, although the information is in the micrographs.
