Explainable AI
While some strcutural biology tasks lend themselves easily to automation, this is not universally true for machine learning applications. Especially those data sets beyond one's own lab to obtain more biological insight are often not trusted and only used frequently if they really provide outstanding new information.
It is challenging for scientists to comprehend how an AI algorithm arrives at a result, effectively making it a black box. This, of course, sits uncomfortably with those who professionally strive for understanding! For many, it makes AI untrustworthy. Therefore, it is crucial to explain how an AI-enabled system has arrived at a specific output. It can help developers ensure that the system is working as expected or facilitate challenging the outcome. It helps to understand the underlying principles – to ask the AI what it has learned and how to describe this knowledge or to explicitly correct it if it learnt something undesirable. In some areas, such as medicine or finance, explainability can also be a moral obligation. This need has spawned the development of several methods that may be transferable to applications in structural biology, collectively known as “explainable AI”.
One approach is the ask which parts of a given input were the most important contributors in the determination of the output. For simpler networks, this can be achieved in the general case. For CNNs, this is called layer wise relevance propagation (LRP) and is a technique for determining which features in a particular input vector contribute most strongly to a neural network's output. An example is shown below:
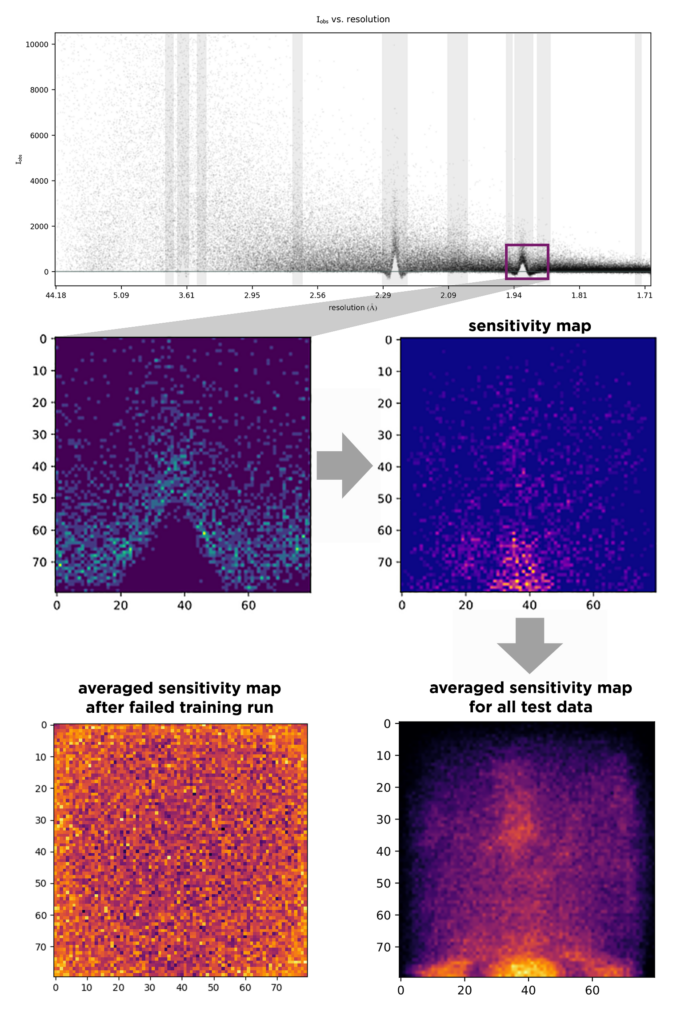
AUSPEX plot of measured reflection intensity in macromolecular crystallography versus resolution. In order to train a neural network to recognize ice rings, which form distinct spikes in the distribution of intensities and resolution, an ice ring range (purple box) is chosen and scaled to 80 by 80 pixels and normalized (middle left). The Helcaraxe convolutional network takes these pictures and annotates them individually as “ice ring” or “no ice ring”. SmoothGrad, an algorithm that identifies regions of an input image that were particularly influential to the final classification, outputs a “sensitivity map”, for both each individual distribution (middle right) or as an average across a whole dataset (bottom right), yielding a visual sanity check of whether the network is “looking at the right things”. In the case of AUSPEX, the region under the spike as well as the regions left and right from the spike are very important for the network’s decision making, as they would be for a human - while on the left, where training did not work (too little training data), the output does not comply with expectation or common sense. During the development of this new method, these sensitivity map allowed the identification of problems and bugs during network training (bottom left).
Related methods allow us to analyse which neurons contributed most strongly, in order to not only follow the path of information through the neural network, but also to permit parametrization of a system with neural network training. This can be facilitated by using a chain of multiple neural networks that each give human-readable outputs. For example, Alphafold1 predicted a distance matrix and used this for a gradient descent algorithm. The matrix and torsion angle distributions that result are totally understandable by humans. In AlphaFold, all information is fed directly into the next network, which might be more efficient, but is also less transparent to users. As a last resort, when no general statements can be made (for example for very complex systems) one distinct output can be explained by tracing it back through the non-linear model that produced it, a so-called “local interpretation”.
As techniques in explainable AI gain more power and significance, they will also be used in structural biology applications, where the findings can then be used to support the development of classical statistical analysis, model parametrization and decision-making algorithms. Full acceptance by the community is likely only to arise from a rational explanation of what the algorithm is doing and, from there, an improved understanding of physical laws of nature.
We have teamed up with collaboration partners in the field, such as the CCP4/CCP-EM machien learning task force, Prof. Nihat Ay and Prof. Philip Kollmannsberger in order to explore and popularize explainable AI in strcutural biology. We also use them in the AUSPEX Verbundforschungsprojekt in order to determine statistical quality indicators from the learning of neural networks.
