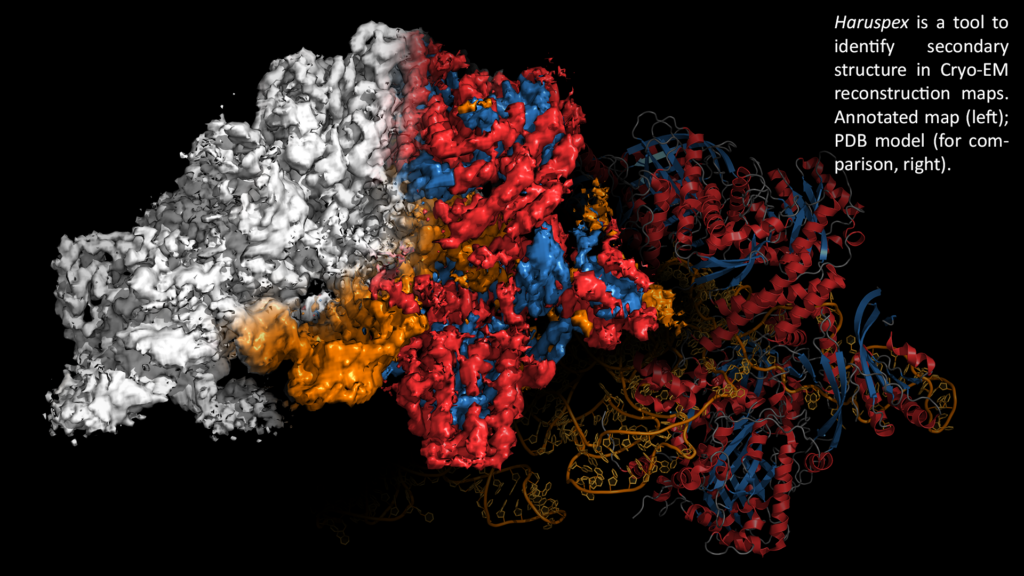
HARUSPEX
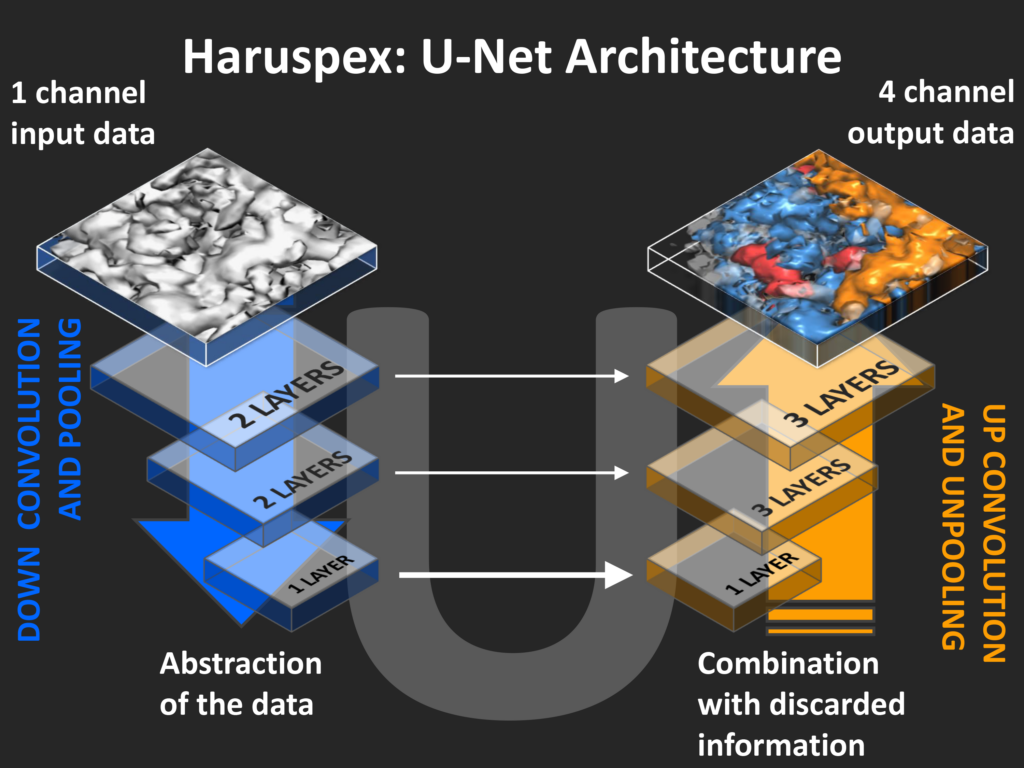
Haruspex is a new tool to identify secondary structure and RNA/DNA in cryo-EM reconstruction maps. It uses a neural network which has been trained on a carefully curated set of 293 experimentally derived reconstructions, called EMDBest (manuscript in preparation) – which is, to our knowledge, currently the largest manually curated training data set of cryo-EM data. Due to its high recall and precision rates of 95.1% and 80.3%, respectively, on an independent test set of 122 maps, Haruspex can also be used for validation during model building. Haruspex is freely available since the start of 2021 as part of the CCP‐EM software suite.
The correct interpretation of lower resolution domains, as well as findings by other groups make it likely that Haruspex, with a suitable training set, could be adapted to lower average reconstruction map resolutions. However, careful work will be needed to find low resolution reconstruction maps for which an accurate model exists which can be used as training data.
More info:
https://www.ccpem.ac.uk/downloads/symposium/thorn_CCP-EM_Haruspex.pdf
Tutorial:
https://www.ccpem.ac.uk/downloads/tutorials/haruspex_slac/haruspex_tutorial.pdf
Haruspex is part of our wider reserach interest to extract and fully exploit cryo-EM. Crystallography has directly impacted how Cryo-EM data are processed and interpreted: Atomic models are fitted to reconstruction maps using restraints and methods which were originally developed for crystallographic structure solution.
While this has been a serviceable interim solution until now, the limitations of this approach have also become clear. The size of the structures alone poses a huge challenge to manual and automatic model building (and computing facilities) alike, with automatic map interpretation only being possible at resolutions better than 3-4 Å. The inherent flexibility and heterogeneity of the sample further limits the resolutions that reconstruction maps can achieve, and our current models cannot adequately account for this heterogeneity, or correctly model radiation damage and charge distribution within the molecular assembly. Finally, the model is fitted to a reconstruction map which, while derived from the data, does not represent the wealth of information contained in single-particle-micrographs. This means much biologically relevant information is lost in the process of reconstruction map calculation, such as information on the correlation of movements. We believe that this can be changed with modern network architectures in such as rotation-invariant tensor field networks, GANs and linguistic models, and are searching for funding partners to realize this technology.